MATLAB Remote Jobs Submission¶
MATLAB Parallel Server is a set of MATLAB functions that extend the capabilities of the MATLAB Parallel Computing toolbox to allow you to submit jobs from directly to a remote HPC cluster.
This allows you to take full advantage of the computing power available on the CRC's clusters to perform complex parallelized computations in MATLAB. Parallel jobs can be submitted either from an SBATCH submission on the clusters or on your own computer with custom a set of custom scripts that integrate with Slurm. In fact, you must use the MATLAB Parallel Server if you want to run MATLAB jobs that span multiple nodes.
When you submit a MATLAB Parallel Server job from your MATLAB desktop client, scripts and data are transferred to the cluster by MATLAB. Results from jobs that run on the cluster are readily available to MATLAB running on your desktop.
The instructions on this page are for people who want to interact with Parallel Server from their own local machine.
Instructions for submitting MATLAB Parallel Server jobs using sbatch scripts on the compute node are available on the
cluster: /ihome/crc/how_to_run/matlab
Local Client Job Submission¶
Prerequisites¶
- Your MATLAB version must be at least R2021a. Plugin scripts are also available for MATLAB 2022a. If you don’t have one of these versions, please visit the Pitt IT Software Download Service, search for "MATLAB" and download and install R2021a or R2022a. Note: The 2022a plugin scripts seem to work fine with previous versions, but will work best with 2022a MATLAB.
- Make sure you are using the newest update of your version of MATLAB (Open MATLAB > "HOME" Tab > "RESOURCES" Section> "Help" Drop Down > Check for Updates)
- You must have the MATLAB Parallel Computing Toolbox installed. To see what toolboxes are
installed, type the
vercommand in the MATLAB command window. You should seeParallel Computing Toolboxlisted. If you don't, you can open MATLAB > go to the home tab > Add-Ons > Get Add-Ons > Search forParallel Computing Toolboxand Install (If you are using MATLAB licensed through the university). - You must be connected to the CRC VPN in order to submit jobs from your desktop, if you are off-campus. Please visit this page on installing the vpn.
Download the Cluster Plugin Scripts¶
In order to submit jobs from your desktop, you will need to install some custom MATLAB plugin scripts that know how to interact with the Slurm job scheduler on a cluster. Use the links below to download the non-shared filesystem (used by your local client) script bundle for your version of MATLAB.
Pitt.h2p.R2022a.zip (recommended)
Note
The instructions below have been updated to correspond to the R2022a plugin scripts. Some cluster object fieldnames have changed. Changes from the previous scripts will have comments next to them.
You should not use the above plugin scripts when running directly on the cluster as they share a filesystem with the MATLAB installation that is running the jobs! If you are running Matlab from the cluster, such as VIZ, then there is no need to download and execute the plugin scripts. In that case, directly run "configCluster" from Matlab to create the cluster profile.
Unpack the Cluster Plugin Bundle to a Path on Your Computer¶
Copy the integration bundle file to a new folder on your computer, such as Downloads/matlab-parallel-server.
Unpack the plugin bundle file into the path you choose.
Run the configCluster.m Script to Create a Cluster Profile¶
Open MATLAB. In the Current Folder window, navigate to the path where you unpacked one of the cluster plugin bundles.
In that path, find the Matlab script called configCluster.m. Run this script by right-clicking on the name and
selecting Run. (If MATLAB asks you if you want to add the path to your environment, select Add Path.)
Submission to the remote cluster requires SSH credentials. The configCluster script will prompt you to enter the
username you use to log in to the cluster. Just enter your Pitt username in lower case (e.g. abc123). You should not
append "@pitt.edu" to it. The script also asks you your group name on H2P cluster. If you do not know what your group
name is, login to the cluster and run the following command:
finger <your Pitt username>
For example:
[shs159@login1 ~]$ finger shs159
Login: shs159 Name: shs159
Directory: /ihome/pgivi/shs159 Shell: /bin/bash
On since Tue Aug 3 15:06 (EDT) on pts/124 from login0.crc.pitt.edu
4 days idle
No mail.
No Plan.
[shs159@login1 ~]$
Here in /ihome/<your group>/<your username>, the first subdirectory is your group name and the second directory one
step further is your Pitt username.
After you enter your username and your group name, the program will display an informational reminder that you will
need to set the NumNodes and WallTime parameters before you can submit a job. We'll cover how to set job parameters
in the next section. A screenshot of a successful run of configCluster is shown below:
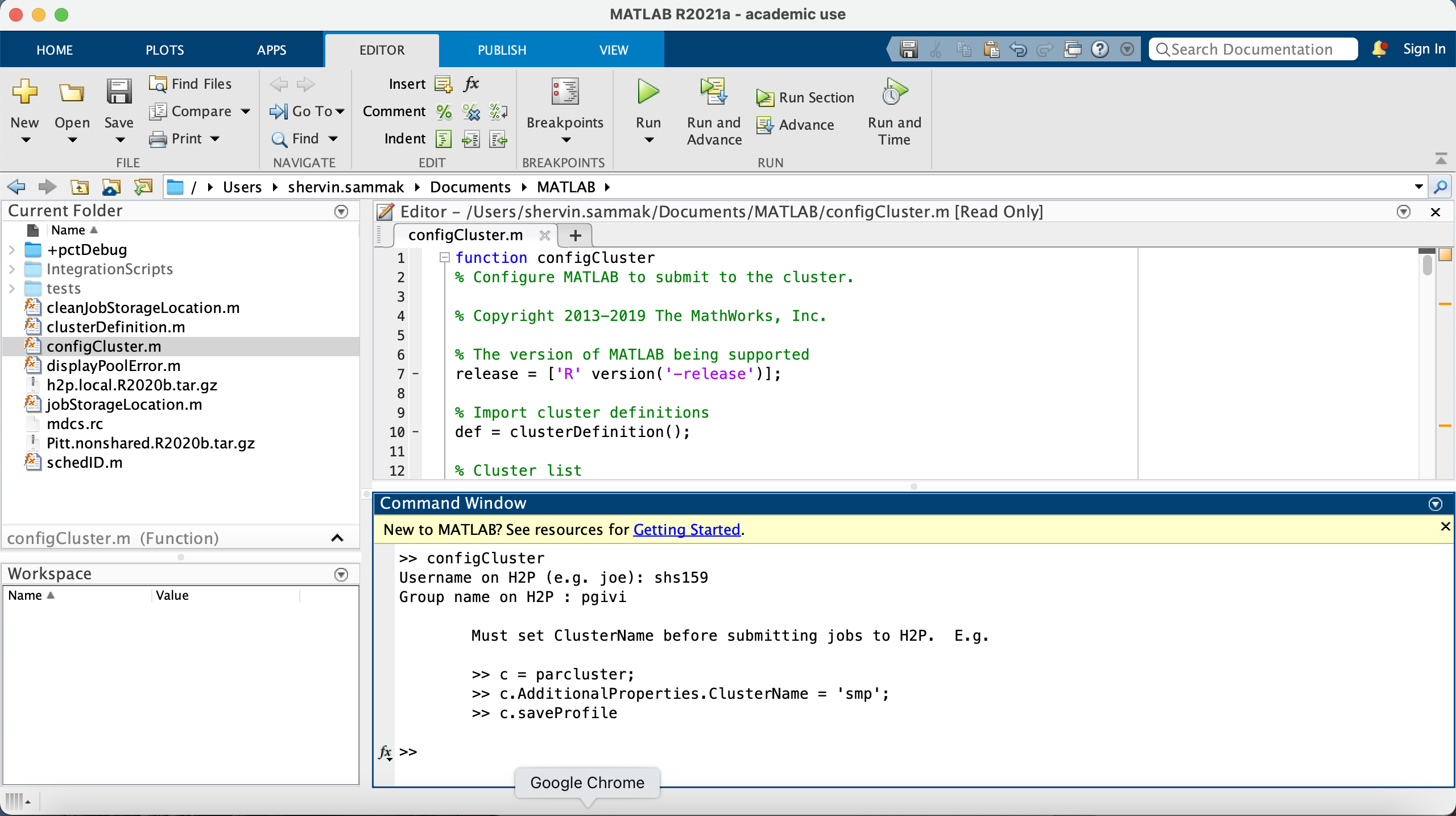
Jobs will now default to the cluster rather than submit to the local machine.
Note
If you would like to submit to the local machine then run the following command:
>> % Get a handle to the local resources
>> c = parcluster('local');
Working with Cluster Objects¶
Using cluster objects is central to using MATLAB Parallel Server. The parcluster command is used to initialize a cluster object for your session:
>> c=parcluster
c =
Generic Cluster
Properties:
Profile: h2p R2022a
Modified: false
Host: localhost
NumWorkers: 100000
NumThreads: 1
JobStorageLocation: /Users/shervin.sammak/MdcsDataLocation/h2p/Shervins-MBP.fios-router.home/R2022a/remote
ClusterMatlabRoot: /ihome/crc/install/matlab/R2022a
OperatingSystem: unix
RequiresOnlineLicensing: false
PluginScriptsLocation: /Users/shervin.sammak/Documents/MATLAB/IntegrationScripts/h2p
AdditionalProperties: List properties
Associated Jobs:
Number Pending: 0
Number Queued: 0
Number Running: 3
Number Finished: 0
>> myjob2 = batch(c,'plot_sine','AutoAddClientPath' ,false, 'CurrentFolder', './test');
3x1 Job array:
ID Type State FinishDateTime Username Tasks
-----------------------------------------------------------------------------------
1 2 independent running shervin.sammak 1
2 3 pool finished shervin.sammak 40
3 4 pool finished shervin.sammak 9
Above, we first created a cluster object c using the 'Pitt h2p R2022a' profile. Then, we entered 'c' without a
semicolon to see the list of properties in the cluster object. Notice that there is a very important subset of
properties called AdditionalProperties. You will need to modify some of these properties values to actually submit a
job to Slurm. Let's examine the AdditionalProperties:
>> c.AdditionalProperties
ans =
AdditionalProperties with properties:
AccountName: ''
AdditionalSubmitArgs: ''
ClusterHost: 'h2p.crc.pitt.edu'
ClusterName: 'smp'
EmailAddress: ''
EnableDebug: 0
Feature: ''
GpusPerNode: 0
MemUsage: ''
Nodes: 0 % Was NumNodes
ProcsPerNode: 0
Partition: '' % Was QueueName
RemoteJobStorageLocation: '/ihome/pgivi/shs159/MdcsDataLocation/h2p/Shervins-MBP.fios-router.home/R2022a/remote'
Reservation: ''
% UseIdentityFile Removed
UseSmpd: 0
Username: 'shs159'
WallTime: ''
>>
As you can see, when you first create the cluster object, many of the properties do not have values. As a minimum, set values for Nodes (Default is 1), ProcsPerNode (Default is 1), ClusterName (must be specified), and WallTime (must be specified). Optionally, you can set the EmailAddress, Partition, and AdditionalSubmitArgs. The AdditionalSubmitArgs is used for passing any additional options to the Slurm sbatch command.
Examples¶
The examples below show how to use MATLAB Parallel Server to run
-
serial (single CPU) jobs
-
multiprocessor jobs on a single machine
-
and distributed parallel jobs running on multiple nodes.
If you'd like to download MATLAB scripts used in the examples, they are available here: tests.zip
Example 1: serial code¶
In the first example, we will run a simple MATLAB job that uses a single processor. This program isn't parallelized,
but we can still execute the job on the cluster. We'll begin by creating a MATLAB script called mywave.m simply
calculates a million points on a sine wave in a for loop. The file mywave.m has been created in our current MATLAB
folder. The function is pretty simple:
%-- mywave.m --%
tic
for i = 1:1000000
A(i) = sin(i*2*pi/102400);
end
toc
When you run this following commands in MATLAB, it should prompt you for your password for logging in to the cluster. Answer “No” to the question and then enter your Pitt password in the following window. MATLAB caches your password during this session, so you don't have to enter your again for submitting other jobs during the same MATLAB session.
>> c = parcluster;
>> c.AdditionalProperties.ClusterName = 'smp';
>> c.AdditionalProperties.WallTime='1-10:00:00'; %% 1 day and 10 hours wall-time.
>> myjob = batch(c,'mywave','AutoAddClientPath',false);
additionalSubmitArgs =
'--ntasks=1 --cpus-per-task=1 --ntasks-per-core=1 --cluster smp'

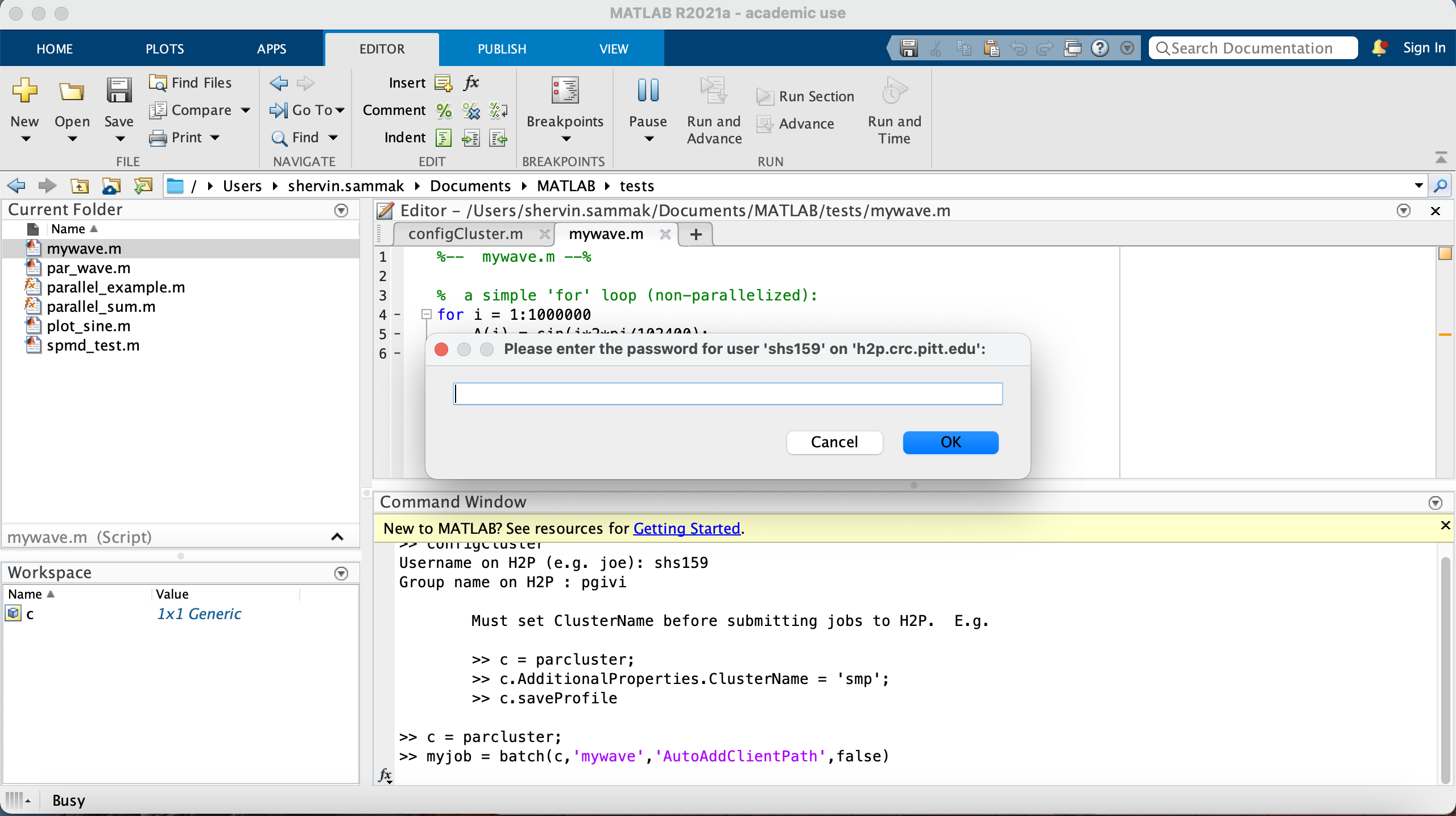
Then, you can monitor and retrieve data:
>> myjob.State
ans =
'finished'
>> myjob.fetchOutputs
ans =
1×1 cell array
{1×1 struct}
>> myjob.load # this command loads all variables from the finished job
>> plot(A)
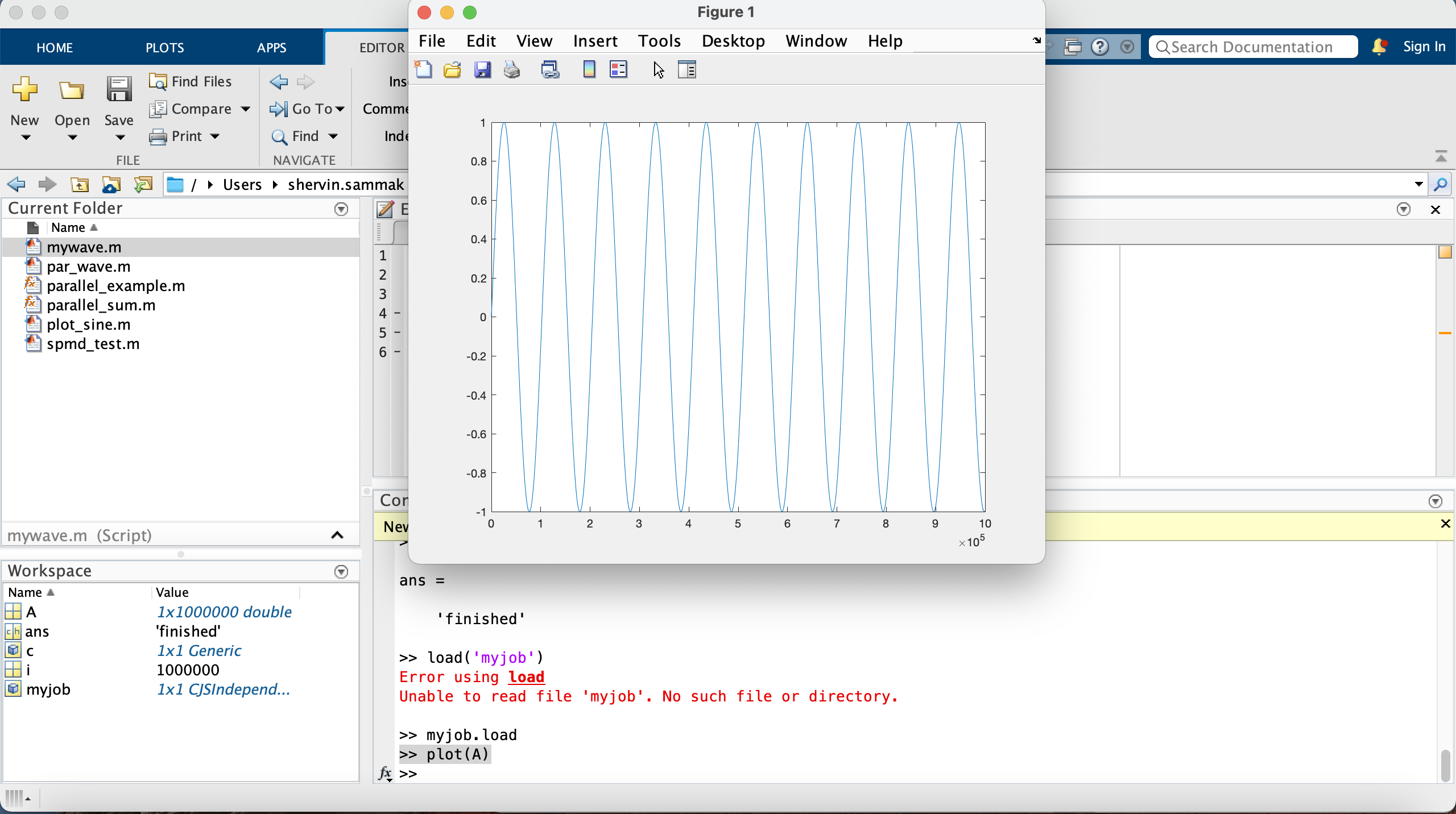
Example 2: Parallel job on a single node¶
In the next example, we will run a parallel job using 8 processors on a single node. We will be using a
parallelized version of mywave.m called par_wave.m that uses the parfor statement to parallelize the previous
for loop:
%-- parallel_mywave --%
% A parfor loop will use parallel workers if available.
tic
parfor i = 1:10000000
A(i) = sin(i*2*pi/2500000);
end
toc
The parfor command will use a pool of workers to execute the loop in parallel if a worker pool exists.
You can learn more about the use of
parfor loops on this page.
Now we'll submit the job to the smp cluster, high-mem partition. Note that the batch command has changed significantly by adding a pool for 7+1 workers. Note that these jobs will always request N+1 CPU cores, since one worker is required to manage the batch job and pool of workers. For example, a job that needs seven workers will consume eight CPU cores.
>> c.AdditionalProperties.ClusterName = 'smp';
>> c.AdditionalProperties.Partition = 'high-mem'; % Was QueueName
>> myjob = batch(c,'par_wave','AutoAddClientPath',false,'Pool', 7);
Example 3: Parallel job on multiple nodes¶
In the next example, we will run a parallel job using 95 + 1 processors across two nodes.
We will be using the same par_wave.m script from Example 2.
MPI jobs require a minimum of 2 Nodes, and must use the full amount of cores that each node offers. For example, a 2 node MATLAB job submitted to the mpi partition (48 cores per node) of the MPI cluster needs to request (2 * 48) - 1 = 95 cores (MATLAB uses 1 worker core to handle background tasks in remote job submission).
>> c.AdditionalProperties.ClusterName = 'mpi';
>> c.AdditionalProperties.Nodes = 2;
>> c.AdditionalProperties.Partition = 'mpi'; % Was QueueName
>> c.AdditionalProperties.WallTime='01:00:00'; %% 1 hour wall-time.
>> myjob = batch(c,'par_wave','AutoAddClientPath',false,'Pool', 95);
additionalSubmitArgs =
'--ntasks=96 --cpus-per-task=1 --ntasks-per-core=1 --cluster mpi -N 2 -p mpi -t 01:00:00'
The above script submits par_wave to the mpi cluster's mpi partition with the total of 39+1 workers.
>> myjob.State
ans =
'finished'
>>
>> myjob.diary
--- Start Diary ---
Elapsed time is 1.297303 seconds.
--- End Diary ---
FAQs¶
Q: My job's .State value is 'failed'. How can I find more information?
A: If a job produces an error, you can call the getDebugLog method to view the error log file. When submitting independent jobs, with multiple tasks, specify the task number.
%Load parcluster
>> c = parcluster
c =
Generic Cluster
Properties:
Profile: h2p R2021a
Modified: false
Host: LAPTOP-H53N4LSB
NumWorkers: 100000
NumThreads: 1
JobStorageLocation: C:\Users\Nicko\AppData\Local\Temp\Nicko\MdcsDataLocation\h2p\LAPTOP-H53N4LSB\R2021a\remote
ClusterMatlabRoot: /ihome/crc/install/matlab/R2021a
OperatingSystem: unix
RequiresOnlineLicensing: false
PluginScriptsLocation: C:\Users\Nicko\Downloads\matlab-parallel-server\Pitt.nonshared.R2021...
AdditionalProperties: List properties
Associated Jobs:
Number Pending: 0
Number Queued: 1
Number Running: 0
Number Finished: 3
%Take a look at your jobs
>> c.findJob
ans =
4x1 Job array:
ID Type State FinishDateTime Username Tasks
-----------------------------------------------------------------------------
1 1 independent failed Nicko 1
2 2 independent failed Nicko 1
3 3 independent queued Nicko 1
4 4 independent failed Nicko 1
%Create a variable for the job you want to view debug output for
>> j = c.findJob('ID',1)
j =
Job
Properties:
ID: 1
Type: independent
Username: Nicko
State: failed
SubmitDateTime: 14-Apr-2022 10:32:35
StartDateTime:
Running Duration: 0 days 0h 0m 0s
NumThreads: 1
AutoAttachFiles: true
Auto Attached Files: List files
AttachedFiles: {}
AutoAddClientPath: false
AdditionalPaths: {}
EnvironmentVariables: {}
Associated Tasks:
Number Pending: 0
Number Running: 0
Number Finished: 1
Task ID of Errors: []
Task ID of Warnings: []
Task Scheduler IDs: 5707953
% View the debug output for the set of tasks being performed by the job.
>> c.getDebugLog(j.Tasks)
LOG FILE OUTPUT:
Executing: /ihome/crc/install/matlab/R2021a/bin/worker
======BEGIN LICENSE MANAGER ERROR======
License checkout failed.
License Manager Error -5
Cannot find a license for MATLAB_Distrib_Comp_Engine.
Troubleshoot this issue by visiting:
https://www.mathworks.com/support/lme/R2021a/5
Diagnostic Information:
Feature: MATLAB_Distrib_Comp_Engine
License path: /ihome/sam/nlc60/.matlab/R2021a_licenses:/ihome/crc/install/matlab/R2021a/licenses/license.dat:/ihom
e/crc/install/matlab/R2021a/licenses/network.lic
Licensing error: -5,147.======END LICENSE MANAGER ERROR======
Exiting with code: 1
Q: If I lose my VPN connection or put my local machine in sleep mode, can I still retrieve my jobs?
A: Yes, this is as simple as calling parcluster again.
>> c=parcluster
c =
Generic Cluster
Properties:
Profile: h2p R2021a
Modified: false
Host: localhost
NumWorkers: 100000
NumThreads: 1
JobStorageLocation: /Users/shervin.sammak/MdcsDataLocation/h2p/Shervins-MBP.fios-router.home/R2021a/remote
ClusterMatlabRoot: /ihome/crc/install/matlab/R2021a
OperatingSystem: unix
RequiresOnlineLicensing: false
PluginScriptsLocation: /Users/shervin.sammak/Documents/MATLAB/IntegrationScripts/h2p
AdditionalProperties: List properties
Associated Jobs:
Number Pending: 0
Number Queued: 0
Number Running: 1
Number Finished: 2
3x1 Job array:
ID Type State FinishDateTime Username Tasks
-----------------------------------------------------------------------------------
1 2 independent running shervin.sammak 1
2 3 pool finished 16-Aug-2021 09:18:04 shervin.sammak 40
3 4 pool finished 16-Aug-2021 09:18:31 shervin.sammak 9
Q: If my MATLAB script has created a file on the cluster, do I need to manually copy the file back onto my local machine?
A: Yes, here's an example which plots a sine function and create a plot called sine_wave.ps:
% Plot the sine wave
disp('This script plots the sine wave');
x = -pi:.1:pi;
y = sin(x);
p = plot(x,y)
set(gca,'XTick',-pi:pi/2:pi)
set(gca,'XTickLabel',{'-pi','-pi/2','0','pi/2','pi'})
xlabel('-\pi \leq \Theta \leq \pi')
ylabel('sin(\Theta)')
title('Plot of sin(\Theta)')
% \Theta appears as a Greek symbol (see String)
% Annotate the point (-pi/4, sin(-pi/4))
text(-pi/4,sin(-pi/4),'\leftarrow sin(-\pi\div4)','HorizontalAlignment','left')
% Change the line color to red and
% set the line width to 2 points
set(p,'Color','red','LineWidth',2)
disp('Now outputing plot to sine_wave.ps');
print -deps sine_wave.ps;
exit;
This is the command to submit the job to the cluster:
myjob2 = batch(c,'plot_sine','AutoAddClientPath' ,false, 'CurrentFolder', './test');
additionalSubmitArgs =
'--ntasks=1 --cpus-per-task=1 --ntasks-per-core=1 --cluster smp -p smp'
The above script submit the plot_sine job to the smp cluster, partition smp where the figure will be saved
in /ihome/<your group>/<your Pitt username>/test.
>> myjob2.diary
Warning: The diary of this batch job might be incomplete because the job is still running.
--- Start Diary ---
This script plots the sine wave
p =
Line with properties:
Color: [0 0.4470 0.7410]
LineStyle: '-'
LineWidth: 0.5000
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [1x63 double]
YData: [1x63 double]
ZData: [1x0 double]
Use GET to show all properties
Now outputing plot to sine_wave.ps
--- End Diary ---
>>
Q: Where are metadata files stored on my local machine?
A: The cluster object has a field called JobStorageLocation that defines this:
JobStorageLocation: C:\Users\Nicko\AppData\Roaming\MathWorks\MATLAB\3p_cluster_jobs\h2p\LAPTOP-H53N4LSB\R2022a\nonshared
You can also access the metadata on the cluster with the cluster.AdditionalProperties.RemoteJobStorageLocation field:
RemoteJobStorageLocation: '/ihome/sam/nlc60/.matlab/3p_cluster_jobs/h2p/LAPTOP-H53N4LSB/R2022a/nonshared'
Please note that if you run configCluster again, all metadata will be lost on your local machine.
Q: I was disconnected from the VPN, and now my job submissions are failing. How do I fix this?
Warning: Failed to remove the remote
files for job 8. You may need to delete
the job files from
/ihome/sam/nlc60/.matlab/3p_cluster_jobs/h2p/LAPTOP-H53N4LSB/R2022a/nonshared
manually. Reason:
Failed to execute command on remote
host.
> In parallel.cluster/RemoteClusterAccess/startMirrorForJob (line 625)
In communicatingSubmitFcn (line 175)
In parallel.cluster/Generic/submitJobCommon (line 689)
In parallel.cluster/Generic/hSubmitCommunicatingJob (line 533)
In parallel.internal.cluster/CJSJobMixin/submitOneJobCommon (line 66)
In parallel.job/CJSCommunicatingJob/submitOneJob (line 88)
In parallel/Job/submit (line 404)
In parallel.internal.cluster/BatchHelper2/doBatch (line 48)
In parallel.Cluster/batch (line 155)
Error using parallel.Cluster/batch
Job submission failed because the plugin
function 'communicatingSubmitFcn.m'
errored.
Caused by:<br />
Error using<br />
parallel.cluster.RemoteClusterAccess/startMirrorForJob<br />
Failed to start mirror for job<br />
with ID 8.<br />
Error using<br />
parallel.internal.remoteaccess.FileMirror/addToMirror<br />
Failed to execute<br />
command on remote host.
A: The command is failing on the remote host because you need to re authorize your connection with your username and password.
Reestablish your cluster object with the parcluster command, and then submit.
Q: My submission to MPI yields the following error: "Your MPI job is not requesting the full number of cores on its nodes: 48 cores on the mpi partition" How do I resolve this?
A: MPI jobs require a minimum of 2 Nodes, and must use the full amount of cores that each node offers. For example, a 2 node MATLAB job submitted to the mpi partition (48 cores per node) of the MPI cluster needs to request (2 * 48) - 1 = 95 cores (MATLAB uses 1 worker core to handle background tasks in remote job submission).
c.AdditionalProperties.Partition = 'mpi'
c.AdditionalProperties.Nodes = 2;
myjob = batch(c,'my_job','AutoAddClientPath', false, 'Pool', 95);
Will send the following job configuration to SBATCH:
'--ntasks=96 --cpus-per-task=1 --ntasks-per-core=1 --cluster mpi -N 2 -p mpi -t 01:00:00'